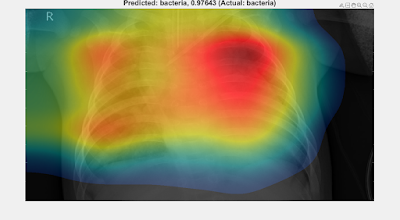
UPDATES: See Part 4 for a grad-CAM analysis of all the trained networks presented below, then Part 5 where the grad-CAM results of Part 4 are used to train another suite of networks to help choose between the lung X-ray classifiers presented below.
*** DISCLAIMER ***
I have no medical training. Nothing presented here should be considered in any way as informative from a medical point-of-view. This is simply an exercise in image analysis via Deep Learning using MATLAB, with lung X-rays as a topical example in these times of COVID-19.
INTRODUCTION
In this Part 3 in my series of blog articles on exploring Deep Learning of lung X-rays using MATLAB, the analysis of Part 1 is revisited but rather than just using the pre-trained googlenet as the basis of Transfer Learning, the performance of all the pre-trained networks available via MATLAB as the basis for the Transfer Learning procedure is compared.
AVAILABLE PRE-TRAINED NETWORKS
See this overview for a list of all the available pre-trained Deep Neural Networks bundled with MATLAB (version R2020a). There are 19 available networks, listed below, which include two alternate versions of googlenet: the original, and the alternate version with identical layer structure but pre-trained on images of places rather than images of objects.
| Available Pre-Trained Networks |
| squeezenet |
| googlenet |
| googlenet (places) |
| inceptionv3 |
| densenet201 |
| mobilenetv2 |
| resnet18 |
| resnet50 |
| resnet101 |
| xception |
| inceptionresnetv2 |
| shufflenet |
| nasnetmobile |
| darknet19 |
| darknet53 |
| alexnet |
| vgg16 |
| vgg19 |
TRANSFER LEARNING
Network Preparation
Each of the above pre-trained networks were prepared for Transfer Learning in the same manner as described in Part 1 (and references therein). This involved replacing the last few layers in each network in preparation for re-training with the lung X-ray images. To determine which layers to replace required identifying the last learning layer (such as a convolution2dLayer) in each network and replacing from that point onwards using new layers with the appropriate number of output classes (e.g., 2 or 4 etc rather than 1000 as per the pre-trained imageNet classes). For convenience, I've collected together the appropriate logic for preparing each of the networks (since the relevant layer names are generally different for the various networks) in the function prepareTransferLearningLayers which you can obtain from my GitHub repository here.
Data Preparation
For each of the Examples 1--4 in Part 1, the training and validation image datasets were prepared as before (from all the underlying images available) with the important additional action: for each Example, the respective datasets were frozen (rather than randomly chosen each time) so that each of the 19 networks could be trained and tested on precisely the same datasets as one another, thereby enabling ready comparison between network performance.
Training Options
The training options for each case were set as follows:
MaxEpochs=1000; % Placeholder, patience will stop well before
miniBatchSize = 10;
numIterationsPerEpoch = floor(numTrainingImages/miniBatchSize);
options = trainingOptions('sgdm',...
'ExecutionEnvironment','multi-gpu', ...% for AWS ec2 p-class VM
'MiniBatchSize',miniBatchSize, 'MaxEpochs',MaxEpochs,...
'InitialLearnRate',1e-4, 'Verbose',false,...
'Plots','none', 'ValidationData',validationImages,...
'ValidationFrequency',numIterationsPerEpoch,...
'ValidationPatience',4);
miniBatchSize = 10;
numIterationsPerEpoch = floor(numTrainingImages/miniBatchSize);
options = trainingOptions('sgdm',...
'ExecutionEnvironment','multi-gpu', ...% for AWS ec2 p-class VM
'MiniBatchSize',miniBatchSize, 'MaxEpochs',MaxEpochs,...
'InitialLearnRate',1e-4, 'Verbose',false,...
'Plots','none', 'ValidationData',validationImages,...
'ValidationFrequency',numIterationsPerEpoch,...
'ValidationPatience',4);
RESULTS
Refer to Part 1 for the motivation and background details pertaining to the following examples.
EXAMPLE 1: "YES / NO" Classification of Pneumonia
The 19 networks were re-trained (via Transfer Learning) on the relevant training dataset for the given example (1280 images, equally balanced across both classes). The following table shows the performance of each trained network when applied to the validation dataset (balanced, 112 each "yes" / "no") and the holdout dataset (unbalanced, 3806 "yes" only). The results are ordered (descending) by (i) Average Accuracy (across both classes), then (ii) Pneumonia Accuracy (i.e., fraction of "yes" correctly diagnosed). The table also included the Missed Pneumonia rate i.e., the percentage of the total validation population that should have been diagnosed "yes" (pneumonia) but which were missed i.e., wrongly diagnosed as "no" (healthy).
| Base network | Validation: Average Accuracy | Validation: Pneumonia Accuracy | Validation: Healthy Accuracy | Validation: Missed Pneumonia | Holdout: Average Accuracy |
| vgg16 | 91% | 88% | 95% | 6% | 86% |
| alexnet | 90% | 86% | 94% | 7% | 85% |
| darknet19 | 88% | 88% | 88% | 6% | 87% |
| darknet53 | 88% | 89% | 87% | 5% | 89% |
| shufflenet | 88% | 84% | 92% | 8% | 84% |
| googlenet | 88% | 83% | 93% | 8% | 84% |
| googlenetplaces | 88% | 89% | 86% | 5% | 87% |
| resnet101 | 88% | 77% | 98% | 12% | 76% |
| nasnetlarge | 87% | 83% | 91% | 8% | 84% |
| resnet50 | 87% | 86% | 88% | 7% | 88% |
| vgg19 | 86% | 90% | 81% | 5% | 91% |
| xception | 86% | 79% | 93% | 11% | 83% |
| resnet18 | 85% | 71% | 100% | 15% | 77% |
| squeezenet | 84% | 92% | 76% | 4% | 91% |
| densenet201 | 83% | 71% | 96% | 15% | 72% |
| inceptionresnetv2 | 83% | 92% | 73% | 4% | 86% |
| nasnetmobile | 72% | 84% | 60% | 8% | 85% |
| inceptionv3 | 72% | 83% | 61% | 8% | 83% |
| mobilenetv2 | 69% | 93% | 46% | 4% | 93% |
EXAMPLE 2: Classification Bacterial or Viral Pneumonia
The 19 networks were re-trained (via Transfer Learning) on the relevant training dataset for the given example (3434 images, equally balanced across both classes). The following table shows the performance of each trained network when applied to the validation dataset (balanced, 320 each "bacteria" / "virus") and the holdout dataset (unbalanced, 520 "bacteria" only). The results are ordered (descending) by (i) Average Accuracy (across both classes), then (ii) Viral Accuracy (i.e., fraction of viral cases correctly diagnosed). Also shown is the Missed Viral rate (i.e., the fraction of the total validation population that should have been diagnosed viral but which were missed (wrongly diagnosed as bacterial).
| Base network | Validation: Average Accuracy | Validation: Viral Accuracy | Validation: Bacterial Accuracy | Validation: Missed Viral | Holdout: Average Accuracy |
| darknet53 | 80% | 76% | 84% | 12% | 84% |
| vgg16 | 80% | 73% | 87% | 14% | 83% |
| squeezenet | 79% | 75% | 83% | 12% | 80% |
| vgg19 | 78% | 79% | 78% | 10% | 78% |
| mobilenetv2 | 78% | 81% | 75% | 9% | 71% |
| googlenetplaces | 78% | 71% | 86% | 15% | 85% |
| densenet201 | 78% | 70% | 87% | 15% | 85% |
| inceptionresnetv2 | 78% | 82% | 74% | 9% | 70% |
| alexnet | 78% | 81% | 75% | 10% | 71% |
| googlenet | 77% | 71% | 83% | 15% | 83% |
| nasnetlarge | 77% | 78% | 76% | 11% | 76% |
| darknet19 | 77% | 62% | 92% | 19% | 89% |
| inceptionv3 | 76% | 91% | 60% | 4% | 58% |
| resnet50 | 75% | 68% | 83% | 16% | 81% |
| nasnetmobile | 74% | 66% | 81% | 17% | 76% |
| shufflenet | 69% | 50% | 89% | 25% | 88% |
| xception | 69% | 43% | 94% | 28% | 90% |
| resnet101 | 65% | 38% | 92% | 31% | 93% |
| resnet18 | 58% | 80% | 35% | 10% | 39% |
EXAMPLE 3: Classification of COVID-19 or Other-Viral
The 19 networks were re-trained (via Transfer Learning) on the relevant training dataset for the given example (130 images, equally balanced across both classes). The following table shows the performance of each trained network when applied to the validation dataset (balanced, 11 each "covid" / "other-viral") and the holdout dataset (unbalanced, 1938 "other-viral" only). The results are ordered (descending) by (i) Average Accuracy (across both classes), then (ii) COVID-19 Accuracy (i.e., fraction of COVID-19 cases correctly diagnosed). Also shown is the Missed COVID-19 i.e., the fraction of the total validation population that should have been diagnosed COVID-19 but which were missed (wrongly diagnosed as Other-Viral).
| Base network | Validation: Average Accuracy | Validation: COVID-19 Accuracy | Validation: Other-Viral Accuracy | Validation: Missed COVID-19 | Holdout: Average Accuracy |
| alexnet | 100% | 100% | 100% | 0% | 95% |
| vgg16 | 100% | 100% | 100% | 0% | 96% |
| vgg19 | 100% | 100% | 100% | 0% | 97% |
| darknet19 | 100% | 100% | 100% | 0% | 93% |
| darknet53 | 100% | 100% | 100% | 0% | 96% |
| densenet201 | 100% | 100% | 100% | 0% | 96% |
| googlenet | 100% | 100% | 100% | 0% | 95% |
| googlenetplaces | 100% | 100% | 100% | 0% | 95% |
| inceptionresnetv2 | 100% | 100% | 100% | 0% | 96% |
| inceptionv3 | 100% | 100% | 100% | 0% | 96% |
| mobilenetv2 | 100% | 100% | 100% | 0% | 95% |
| resnet18 | 100% | 100% | 100% | 0% | 96% |
| resnet50 | 100% | 100% | 100% | 0% | 96% |
| resnet101 | 100% | 100% | 100% | 0% | 96% |
| shufflenet | 100% | 100% | 100% | 0% | 95% |
| squeezenet | 100% | 100% | 100% | 0% | 94% |
| xception | 100% | 100% | 94% | 0% | 96% |
| nasnetmobile | 95% | 100% | 91% | 0% | 94% |
| nasnetlarge | 95% | 91% | 100% | 5% | 96% |
EXAMPLE 4: Determine if COVID-19 pneumonia versus Healthy, Bacterial, or non-COVID viral pneumonia
The 19 networks were re-trained (via Transfer Learning) on the relevant training dataset for the given example (260 images, equally balanced across all four classes). The following table shows the performance of each trained network when applied to the validation dataset (balanced, 11 each of "covid" / "other-viral" / "bacterial" / "healthy") and the holdout dataset (unbalanced, zero "covid", 1934 "other-viral", 2463 "bacterial", 676 "healthy"). For succinctness, not all four classes are shown in the table (just the key ones of interest which the network should ideally distinguish: COVID-19 and Healthy). The results are ordered (descending) by (i) Average Accuracy (across all four classes), then (ii) COVID-19 (i.e., fraction of COVID-19 cases correctly diagnosed). Also shown is the Missed COVID-19 i.e., the fraction of the total validation population that should have been diagnosed COVID-19 but which were missed (wrongly diagnosed as belonging to one of the other three classes).
| Base network | Validation: Average Accuracy | Validation: COVID-19 Accuracy | Validation: Healthy Accuracy | Validation: Missed COVID-19 | Holdout: Average Accuracy |
| alexnet | 82% | 100% | 100% | 0% | 58% |
| inceptionresnetv2 | 80% | 100% | 100% | 0% | 61% |
| googlenet | 80% | 91% | 100% | 2% | 61% |
| xception | 77% | 100% | 100% | 0% | 58% |
| inceptionv3 | 77% | 91% | 100% | 2% | 58% |
| mobilenetv2 | 77% | 91% | 100% | 2% | 61% |
| densenet201 | 75% | 100% | 100% | 0% | 61% |
| darknet19 | 75% | 100% | 100% | 0% | 59% |
| nasnetlarge | 75% | 91% | 100% | 2% | 61% |
| vgg19 | 73% | 100% | 100% | 0% | 52% |
| nasnetmobile | 73% | 91% | 100% | 2% | 58% |
| darknet53 | 73% | 91% | 100% | 2% | 63% |
| vgg16 | 73% | 91% | 100% | 2% | 61% |
| googlenetplaces | 73% | 82% | 100% | 5% | 57% |
| resnet18 | 73% | 73% | 100% | 7% | 60% |
| resnet50 | 70% | 91% | 100% | 2% | 61% |
| squeezenet | 70% | 73% | 100% | 7% | 55% |
| shufflenet | 68% | 91% | 91% | 2% | 59% |
| resnet101 | 52% | 64% | 100% | 9% | 51% |
DISCUSSION & CONCLUSIONS
The main points of discussion surrounding these experiments are summarised as follows:
- It is interesting to observe that the best performing networks (i.e., those near the top of the lists of results presented above) per Experiment generally differ per Experiment. The differences must be due to the nature and number of images being compared in a given Experiment and in the detailed structure of the networks and their specific response to the respective image sets in training.
- For each Experiment, the most accurate network turned out not to be googlenet as used exclusively in Part 1. This emphasises the importance of trying different networks for a given problem -- and it is not at all clear a priori which network is going to perform best. The results also suggest that resnet50, as used here, is not actually the optimal choice when analysing these lung images via Transfer Learning.
- Since each Example reveals a different preferred network, a useful strategy for diagnosing COVID-19 could be as follows: (i) use a preferred network from Example 1 (e.g., vgg16 at the top of the list, or some other network from near the top of the list) to determine whether a given X-ray-image-under-test is healthy or unhealthy; (ii) if unhealthy, use a preferred network from Example 2 to determine if viral or bacterial pneumonia; (iii) if viral, use a preferred network from Example 3 to determine if COVID-19 or another type of viral pneumonia; (iv) test the same image using a preferred network from Example 4 (which directly assesses whether-or-not COVID-19). Compare the conclusion of step (iv) with that of step (iii) to see if they reinforce one another by being in agreement on a COVID-19 diagnosis (or not, as the case may be). This multi-network cascaded approach should be more robust than just using a single network (e.g., as per Example 4 alone) to perform the diagnosis.
- Care was taken to ensure that the training and validation sets used throughout were chosen to be balanced i.e., with equal distribution across all classes in the given Experiment. This left the holdout sets i.e., those containing the unused images from the total available pool, comprising an unbalanced set of test images per Experiment representing a further useful test set. Despite the imbalances, the performance on the networks when applied to the holdout images was generally good, suggesting that the trained networks behave consistently.
POTENTIAL NEXT STEPS
- In the interests of time, the training runs were only conducted once per model per Experiment i.e., using one sample of training and validation images per Experiment. For completeness, the training should be repeated with different randomly selected training & validation images (from the available pool) to ensure that the results (in terms of assessing favoured models per Experiment, etc) are statistically significant.
- Likewise, in the interests of time, the training options (hyper-parameter settings) were fixed (based on quick trial-and-error tests, then frozen for all ensuing experiments). Ideally, these should be optimised, for example using Bayesian Optimisation as described here.
- It would be interesting to gain an understanding of the differences in the performance of the various networks across the various Experiments. Perhaps a comparative Activation Mapping Analysis (akin to that presented in Part 2) could shed some light (?)
- It would be interesting to compare the performance of the networks presented in this article with the COVID-Net custom network. Unfortunately, after spending many hours in TensorFlow, I was unable to export the COVID-Net -- either as a Keras model or in ONNX format -- in a manner suitable for importing into MATLAB (via importKerasNetwork or importONNXNetwork). Perhaps, then, the COVID-Net would need to built from scratch within MATLAB in order to perform the desired comparison. I'm not sure if that is possible (given the underlying structure of COVID-Net). Note: I was able to import and work with the COVID-Net model from here in TensorFlow, but could not successfully export it for use within MATLAB.
- Re-train and compare all the models with larger image datasets whenever they become available. If you have access to such images, please consider posting them to the open source COVID-Net archive here.